Knitting is a powerful feature in R that allows you to easily transform your data analysis and visualization into a professional and shareable document. One of the most common output formats for knitting in R is a PDF file. In this step-by-step guide, we will walk you through the process of knitting to PDF in R, so you can create beautiful and organized reports or presentations.
First, you will need to have RStudio installed on your computer. RStudio is an integrated development environment for R that provides a user-friendly interface for writing and running R code. If you haven’t installed RStudio yet, you can download it from the official RStudio website.
Once you have RStudio installed, you will need to open it and create a new R Markdown document. An R Markdown document is a plain text file that combines markdown syntax (which allows you to format text) with R code chunks (which allow you to insert and run R code). To create a new R Markdown document, go to File -> New File -> R Markdown.
After creating the R Markdown document, you can start writing your report or presentation using markdown syntax. Markdown is a lightweight markup language that allows you to format text using simple and intuitive syntax. For example, you can use two asterisks around a word or phrase to make it bold, or use underscores to make it italic. You can also create bullets or numbered lists using hyphens or numbers respectively.
Now it’s time to insert R code chunks into your R Markdown document. R code chunks are enclosed by three backticks followed by the letter “R” and another three backticks. Inside the R code chunks, you can write and run R code. You can use R code to load data, perform calculations, create plots, and more. The output of the R code will be displayed in the final knitted document.
Why knitting to PDF in R is important
R is a powerful programming language commonly used for data analysis and visualization. One of the many features of R is the ability to knit or convert R code and output into various document formats, including PDF.
There are several reasons why knitting to PDF in R is important:
- Shareability: PDF is a widely used document format that can be easily shared across different devices and operating systems. By knitting R code and output to PDF, you can create portable and professional-looking documents that can be viewed and accessed by others.
- Reproducibility: Knitting to PDF allows you to create reproducible reports that include both the code and the output. This is especially useful in data analysis, as it enables others to replicate your work and verify the results. It also makes it easier for you to revisit and reproduce your own analysis in the future.
- Documentation: PDF documents are a popular choice for documentation purposes. By knitting your R code and output to PDF, you can create comprehensive and well-structured reports that provide an overview of your analysis, including the steps taken, the data used, and the results obtained.
- Presentation: PDF documents offer a professional and polished presentation format. By knitting your R code and output to PDF, you can create visually appealing reports with customized formatting, including headers, footers, tables, and images. This allows you to effectively communicate your analysis and findings to others.
- Integration: Knitting to PDF in R seamlessly integrates code, output, and narrative. This means that you can include not only the final results, but also the code that generated them, the plots or tables that illustrate them, and the explanations or interpretations that accompany them. This integration enhances the readability and understanding of your analysis.
In conclusion, knitting to PDF in R is important because it enables you to create shareable, reproducible, well-documented, visually appealing, and integrated reports of your analysis. Whether you are collaborating with others, presenting your work, or simply organizing and documenting your own analysis, knitting to PDF in R is a valuable tool in your data science toolkit.
Step 1: Install the necessary packages
Before we can start knitting to PDF in R, we need to install the required packages. There are a couple of packages that we’ll need to install, including “knitr” and “rmarkdown”.
To install these packages, we can use the “install.packages()” function in R. Open your R console and run the following commands:
- install.packages(“knitr”)
- install.packages(“rmarkdown”)
This will download and install the necessary packages onto your system. Make sure you have an active internet connection for this step.
Once the packages are installed, we can load them into our R session using the “library()” function. Run the following commands to load the packages:
- library(knitr)
- library(rmarkdown)
Now we’re ready to move on to the next step!
Step 2: Load the data and tidy it up
After installing the necessary packages, the next step is to load the data into R and tidy it up for further analysis. In this step, we will cover how to load data from a file and perform some basic data cleaning and manipulation.
1. Load the data:
To load the data into R, we can use the read_csv() function from the tidyverse package. Make sure the file is in the correct directory or provide the file’s full path.
Note: If the data is in a different format, such as Excel or SPSS, you may need to use a different function to read the data.
library(tidyverse)
data <- read_csv("data.csv")
2. Preview the data:
Before performing any further analysis, it’s always a good idea to preview the loaded data to ensure it was read correctly and to get an idea of its structure and contents. The head() function can be used to display the first few rows of the data.
head(data)
3. Clean and tidy the data:
Depending on the nature of the data, it may require some cleaning and tidying before proceeding with the analysis. Common data cleaning tasks include removing duplicate rows, handling missing values, renaming variables, and transforming data types.
In this step, we will focus on a few basic tasks:
- Remove unnecessary variables: If the data contains variables that are not relevant for the analysis, it’s a good practice to remove them. The
select()function can be used to specify which variables to keep or drop. - Rename variables: If the variable names are not descriptive or need to be changed, the
rename()function can be used. - Handle missing values: Missing values can be problematic for many analyses. The
drop_na()function can be used to remove rows with missing values, while themutate()function can be used to replace missing values with a specific value.
data_cleaned <- select(data, -unwanted_variable)
data_cleaned <- rename(data_cleaned, new_variable_name = old_variable_name)
data_cleaned <- drop_na(data_cleaned)
data_cleaned <- mutate(data_cleaned, variable = if_else(is.na(variable), replacement_value, variable))
4. Save the cleaned data:
After cleaning the data, it’s a good practice to save it as a separate file for future use. The write_csv() function can be used to save the cleaned data as a CSV file.
write_csv(data_cleaned, "cleaned_data.csv")
By following these steps, you have successfully loaded the data into R, previewed it, cleaned and tidied it, and saved it as a separate file. The cleaned data is now ready for further analysis and visualization.
Step 3: Create visualizations to include in the final report
Once you have gathered and cleaned your data, it’s time to create visualizations to include in your final report. Visualizations can help to provide a clear and concise representation of your findings, making it easier for others to understand the information you are presenting. In R, there are several packages that you can use to create a wide range of visualizations, including ggplot2, plotly, and lattice.
1. Decide what type of visualization to use:
Before creating your visualizations, it’s important to think about what type of information you want to convey. Do you want to compare different groups or categories? Do you want to show trends over time? Once you have a clear understanding of your goals, you can choose the appropriate visualization technique.
2. Load the necessary packages:
Before you start creating your visualizations, you need to make sure that you have the necessary packages installed and loaded. In R, you can use the install.packages() function to install the packages and then the library() function to load them.
3. Create the visualizations:
Once you have the necessary packages loaded, you can start creating your visualizations. Depending on the type of visualization you want to create, you will use different functions and arguments. For example, if you want to create a bar chart, you can use the ggplot() function from the ggplot2 package.
4. Customize the visualizations:
After creating the basic visualization, you may want to customize it to better present your data. You can add titles, axes labels, legends, and customize the colors, fonts, and styles of the elements. This can be done using different functions and arguments depending on the package you are using.
5. Save the visualizations:
Once you are satisfied with your visualizations, you can save them as image files to include them in your final report. In R, you can use the ggsave() function from the ggplot2 package to save your visualizations as PDF files. You can specify the file name, dimensions, and resolution.
6. Include the visualizations in your final report:
Finally, you can include the visualizations in your final report. This can be done by importing the saved PDF files into your document using a word processor or a document markup language like LaTeX or Markdown. Make sure to provide appropriate captions and references for each visualization.
By following these steps, you can create compelling visualizations to enhance your final report and effectively communicate your findings to others.
Step 4: Generate the final PDF report
Once you have completed all the necessary steps to prepare your data and create the visualizations, it’s time to generate the final PDF report. In this step, we will use the knitr package to convert our R Markdown document into a PDF file.
1. Open the R Markdown file:
Begin by opening the R Markdown file (.Rmd) that you created in Step 3. You can do this in the RStudio editor or any text editor of your choice.
2. Set the output format:
In the first YAML section of your R Markdown file, specify the output format as “pdf_document”. This can be done by adding the following line:
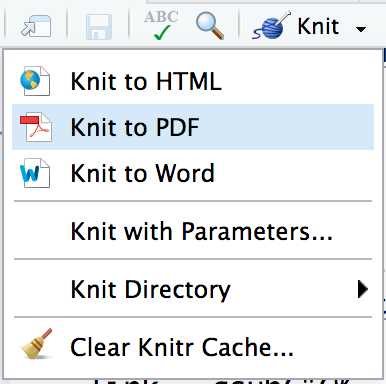
output: pdf_document3. Knit the R Markdown file:
In RStudio, click on the “Knit” button to compile the R Markdown file. This will execute the code chunks, insert the output into the document, and create a PDF file.
4. Review the PDF report:
Once the knitting process is complete, a PDF file with the same name as your R Markdown file will be generated. Open the PDF file to review the final report. Make sure to check the formatting, layout, and content to ensure everything looks as expected.
5. Make adjustments if needed:
If you notice any issues or errors in the PDF report, go back to your R Markdown file and make the necessary adjustments. You can then repeat the knitting process to generate an updated PDF file.
6. Share and distribute the PDF report:
Once you are satisfied with the final PDF report, you can share it with others or distribute it as needed. The PDF format is widely supported and can be easily viewed on different devices and platforms.
That’s it! By following these steps, you can generate a professional-looking PDF report with your knitted R Markdown document.
Troubleshooting common issues
In the process of knitting to PDF in R, you may encounter some common issues. Here are a few troubleshooting tips to help you resolve these problems:
- Package Dependencies: Make sure you have all the necessary packages installed and loaded before knitting to PDF. If you are missing a required package, use the
install.packages()function to install it. - Chunk Execution: Check that all code chunks in your R Markdown document are executing properly. If you encounter errors or unexpected behavior, carefully review the code within the problematic chunk, as well as any preceding or following chunks that may affect it.
- File Paths: Verify that any file paths used in your R Markdown document are correct and accessible. If a file is not found or cannot be read, it will cause issues when knitting to PDF.
- Image Rendering: If you are including images in your R Markdown document, ensure that the file paths to the images are correct. Also, check the image file types supported by the output format you are knitting to. Some formats may require converting images to a specific file type.
- Custom Formatting: If you have applied custom formatting or styling in your R Markdown document, such as font changes or column widths, double-check that the formatting is rendering correctly in the PDF output. If not, review your code and make necessary adjustments.
If you are still experiencing issues after troubleshooting, refer to the documentation or online resources for the specific packages or functions you are using. You can also seek help from the R community through forums, mailing lists, or social media groups.
Remember, troubleshooting is a normal part of the knitting process, and with some perseverance and attention to detail, you can overcome any obstacles and successfully knit your R Markdown document to PDF.
FAQ:
What is the purpose of knitting to PDF in R?
Knitting to PDF in R allows you to save your R code and outputs, such as data tables, plots, and statistical summaries, into a PDF document. This is useful for generating reports or documents that can be easily shared or printed.
How can I knit to PDF in R?
To knit to PDF in R, you can use the ‘knitr’ package. First, you need to install the package by running install.packages(“knitr”). Then, you can use the ‘knit’ function to knit your R code to a PDF document. Simply specify the input file, output file, and desired output format (in this case, “pdf”). For example, you can use the following code: knit(“input.Rmd”, output = “output.pdf”).
Can I customize the appearance of the PDF document when knitting in R?
Yes, you can customize the appearance of the PDF document when knitting in R. The ‘knitr’ package allows you to modify various settings, such as the page size, margins, font size, and table formatting. You can use options like ‘fig.width’ and ‘fig.height’ to control the size of figures, and ‘theme’ to change the overall styling of the document. Additionally, you can use LaTeX commands within your R code chunks to further customize the appearance.
How do I include code and output in the PDF document when knitting in R?
You can include code and output in the PDF document by using R code chunks and inline code within your R Markdown file. R code chunks are denoted by three backticks with the letter ‘r’ immediately following, and are ended with three backticks. Within these code chunks, you can write your R code, and the output will be automatically included in the PDF document. You can also use inline code by enclosing your R code within backticks. For example, \`r 2 + 2\` will be displayed as 4 in the PDF document.
What other output formats can I knit to in R?
In addition to PDF, you can knit your R code to various other output formats, such as HTML, Word, and PowerPoint. The ‘knitr’ package supports multiple output formats, and you can specify the desired output format when using the ‘knit’ function. For example, to knit to HTML, you can use knit(“input.Rmd”, output = “output.html”).
Is there a way to automatically update the PDF document when the underlying R code changes?
Yes, you can automatically update the PDF document when the underlying R code changes by using the ‘watch’ option in the ‘knit’ function. This option allows you to monitor the input file for changes and automatically rebuild the document whenever a change is detected. For example, you can use the following code: knit(“input.Rmd”, output = “output.pdf”, watch = TRUE). This can be especially useful when you are working on a dynamic or iterative analysis and want to keep the PDF document up to date.